Cross-Correlations with halomod¶
The cornerstone of halomod is the TracerHaloModel, which defines the behaviour of dark matter halos and one specific tracer within them. If you want to cross-correlate different tracers, extracting different quantities from two different halo models and calculate the cross-correlation, you can use CrossCorrelations framework. This tutorial goes through a simple example of such a calculation.
[1]:
import numpy as np
from halomod.cross_correlations import CrossCorrelations,_HODCross, ConstantCorr
import matplotlib.pyplot as plt
import halomod
[2]:
print(f"Using halomod v{halomod.__version__}")
Using halomod v1.6.1.dev3+ge514be1.d20200819
A CrossCorrelations object essentially needs three inputs: halo models for two different tracers, and a cross_hod_model to specify how these two tracers cross-correlate, which typically should be constructed by users themselves. Let’s first use a very basic ConstantCorr relation (we’ll go back to what it is later):
[3]:
cross = CrossCorrelations(
cross_hod_model=ConstantCorr,
halo_model_1_params={'z': 1},
halo_model_2_params={'z': 0}
)
This defines the cross-correlation between the same galaxy distribution (the default Zehavi05) at z=1.0 and z=0.0.
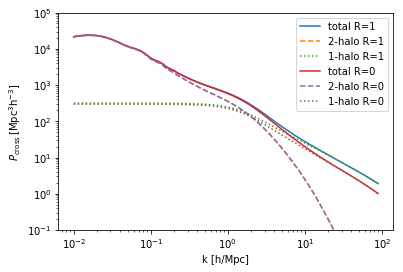
The two halo models within the cross-correlation can be easily extracted and manipulated as they are just TracerHaloModel objects. For example, if we want to change the redshift of the cross-correlation:
[4]:
print(cross.halo_model_1.z)
cross.halo_model_1.z = 0.5
print(cross.halo_model_1.z)
1.0
0.5
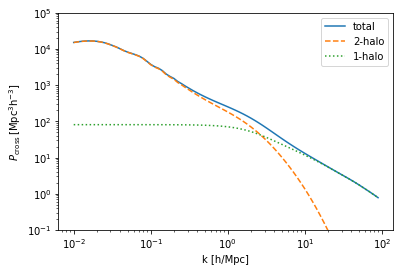
The various two-point statistics of this cross-correlation:
[5]:
plt.plot(cross.halo_model_1.r,cross.corr_cross-1, ls='-', label='total')
plt.plot(cross.halo_model_1.r,cross.corr_2h_cross, ls='--', label='2-halo')
plt.plot(cross.halo_model_1.r,cross.corr_1h_cross, ls=':', label='1-halo')
plt.xscale('log')
plt.yscale('log')
plt.ylim(1e-5,1e5)
plt.legend()
plt.ylabel(r'$\xi_{\rm cross}$')
plt.xlabel(r'r [Mpc/h]');
/home/steven/Documents/Projects/halos/HMF/hmf/src/hmf/density_field/halofit.py:106: UserWarning: sigma_8 is not used any more, and will be removed in v4
warnings.warn("sigma_8 is not used any more, and will be removed in v4")
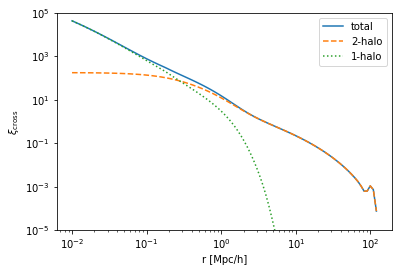
[6]:
plt.plot(cross.halo_model_1.k_hm,cross.power_cross, ls='-', label='total')
plt.plot(cross.halo_model_1.k_hm,cross.power_2h_cross, ls='--', label='2-halo')
plt.plot(cross.halo_model_1.k_hm,cross.power_1h_cross, ls=':', label='1-halo')
plt.xscale('log')
plt.yscale('log')
plt.ylim(1e-1,1e5)
plt.legend()
plt.ylabel(r'$P_{\rm cross}\,[{\rm Mpc^3 h^{-3}}]$')
plt.xlabel(r'k [h/Mpc]');